this article was initially intended to be a quick write-up of something that is all over the internet and sourced pretty much everywhere in a way that i found lacking and/or confusing. what i wanted to do was walk through the information on celiac snps and rsids that is present in a number of places online and present it in a way that was easier to really understand, for those that wanted to check up on it, and not merely take the various websites on authority. what i learned was that the common analysis by the various websites is actually incomplete and that there isn't actually a systematic way to rule out the remaining possible combinations, which actually aren't even all that obscure. i was able to convince myself i cannot develop celiac as an immune response to gluten, but i cannot provide a general algorithm for it, i can only show you how i did it for my own genes and hope it gives you some clues as to how to do it for yours. this article should consequently serve to show you how far you can go with the (flawed) systematic analysis and what is left for you to check on your own.
let's start with the information floating around online, first. the best sites i was able to find presented a three-part test:
1) rule out HLA-DQ2.5 by checking that rs2187668=C,C.
2) rule out HLA-DQ8 by checking for rs7454108=T,T.
3) rule out HLA-DQ2.2 by checking for rs7775228=T,T. if you don't have this specific allele, there are two other ways to rule out hla-dq2.2, instead.
the consensus is that ruling these three genes out rules out the possibility of developing celiac as an immune response to gluten consumption. so, i quickly checked these three steps, got positive results for all three and decided i couldn't have celiac as the cause of my iron deficiency. but, the logic was a little blurry to me - i didn't feel i understood it well enough to be certain, so it kept nagging at me, and i kept running searches related to it. eventually, i decided that i wanted to understand it better before i was sure.
unfortunately, i learned that the the first snp only rules out hla-dq2.5 in cis and does not rule out hla-dq2.5 in trans. what that means is that having that snp can prove you don't have hla-dq2.5 on either of your sixth chromosomes but can't rule out the possibility that you may develop hla-dq2.5 via your two sixth chromosomes meeting in a fuse. by the nature of meiosis, this is always a distinct possibility, if you have half hla-dq2.5 on one chromosome and half on the other. this information exists in the source (often implicitly), but not in the websites citing the source, so it's the websites that are incomplete and not the source that is flawed. it follows that checking this rsid is not enough to prove you don't have hla-dq2.5, and that my exercise in source verification has proven worthwhile in turning up a problem.
the likelihood of this trans pair occurring and it being meaningful in terms of expression is up for debate in the literature, but it can never be ruled out, if you have the constituent parts. if you want to be sure, you're left with the need to disprove the possibility of hla-dq2.5 in trans, as well, and i simply couldn't find a general algorithm to do that.
we can derive an abstract algorithm, but you have to work it out for yourself, concretely. if you have the allele of rs2187668=C,C, and consequently do not have hla-dq2.5 on either copy of your sixth chromosome, there's two ways to go about proving you can't build it in trans, either - you could prove you don't have the alleles required to build hla-dq2.5 in trans, or you could demonstrate that you do have entirely different alleles, and consequently don't have the ones required to build hla-dq2.5 in trans.
for the first option, note that hla-dq2.5 is composed of hla-dqa1*0501 linked to hla-dqb1*0201, primarily. it is important to specify "primarily" because hla-dqa1*0502 may only differ from hla-dqa1*0501 by a few proteins and etc, so very similar linkages could create a gene that behaves essentially identically to hla-dq2.5. for that reason, proving the negative becomes somewhat difficult, as there are quite a few number of possibilities to rule out. as hla-dq2.5 in trans in it's fullest generality would be some kind of hla-dqa1*05 on one of the sixth chromosomes and some kind of hla-dqb1*02 on the other, proving that you do not have this combination means proving that you cannot build that link across the chromosomes, which means proving that you either have neither on either chromosome or, if you have one on one chromosome, then you do not have the other on the other one. i will not be exploring that, here.
i would rather advise the second option, which is demonstrating that you have some other allele instead. i turned out to be homozygous in hla-dq6, and do not have dqa1*05 or dqb1*02 on either of my sixth chromosomes, meaning i cannot build any of the genes of concern in trans linkages.
these are the specific tests i used to demonstrate that, and what they mean:
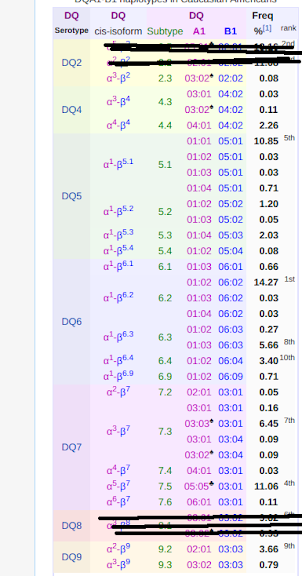
1) rs3135388-T & rs14004-C is a two-step tagging snp for hla-drb1*1501-dqa1*0102-dqb1*0602, which is hla-dq6.2.
2) rs285880-T & rs2273017-T is a two-step tagging snp for hla-drb1*1502-dqa1*0103-dqb1*0601, which is hla-dq6.1.
so, i have hla-dq6.2 in cis on one of my 6th chromosomes and hla-dq6.1 on the other 6th chromosome. that means that my possible trans pairings are dqa1*0102-dqb1*0601 and dqa1*0103-dqb1*0602, both of which are still hla-dq6. it's unavoidable, then - i'm hla-dq6.
i will now show you how i figured that out. i'm presenting this in the form of a stream-of-consciousness discovery, rather than in the form of a reference article. we're going to work this out, together, so there will be many mistakes as i go and i will correct them as they appear. you'll have to read through the whole thing for me to get to the right answer, in the end.
i actually think this is a proof of concept that would be very useful in science education and not something to frown upon. this is how humans actually learn - we make mistakes and correct them. a reference text that consists of the sample space of possible errors is of far more functional utility than one that merely states the right answer.
so, here we go.
how to prove that you don't have celiac using raw dna and make the hipsters cry in their gluten-free beer
sept 30, 2021
as it turns out, the celiac results were easy to find.
HLA-DQ2.5:
rs2187668 6 32605884 C C <------this is immunity for celiac in hla-dq2.5
HLA-DQ8
rs7454108 6 32681483 T T <-------this is immunity for celiac in hla-dq8
celiac happens when an immune response occurs as a result of a mutation in one of these two genes.
no mutation, no celiac.
12:51
oct 1, 2021
ok, i can't sort through 675000 rsids, most of which have weak correlations associated with them.
i need some kind of list of actual known gene locations to run it through.
it seems as though lupus, rheumatoid arthritis and celiac are controlled by a lot of the same genes, though. i tested negative for the first two.
the problem here is that a celiac test is $300 and i'm really not interested in paying for it.
1:47
oct 3, 2021
so, i got my promethease report.
it does not pull out the celiac genes. at all. curiously. it pulls out secondary risk factors, but those shouldn't matter. i've double-checked this and find the information online to be annoyingly vague in it's discussion of "certain alleles" of the hla genes. tell me the alleles, you fuckfaces.
i found this:
unfortunately, the source material doesn't support the claim, unless it's speaking in code.
Primary SNP:
HLA-DQA1/B1 rs2187668
‘C’ = not associated with celiac disease
‘T’ = associated with significantly higher rates of celiac disease
Population Frequency: Around 20% of European descendants carry the “T” allele; its frequency in other populations is 10-15%.
Other Important SNPs:
HLA-DQA1/B1 rs7454108
‘T’ = not associated with celiac disease
‘C’ = associated with higher rates of celiac disease
this is consistent, but i'm running around in circles in terms of finding a source.
the source talks about cis/trans and flips it over, and not about dna base pairs, c and t. i hope somebody didn't get confused.
4:27
this one works out the obscure risk factor, and i don't qualify for that one, either:
but, the sources are still...they don't have the answers i seek.
5:28
i think that what's going on here is that these studies are pointing to alleles in the form of gene variants, and i'm looking for dna base pairs. there's a secret mapping that a professional would know and i need to find.
5:43
HLA-DQ2.5:
rs2187668: C C, which i think is a negative result.
HLA-DQ8
rs7454108: T T, which i think is a negative result
HLA-DQ2.2:
rs4713586 A A, which is a risk factor in the presence of two further [C is a negative factor, here]
rs2395182 T G, which is a risk factor in the presence of two further [the risk factor is T]
rs7775228 T T, which is not a risk factor (the risk factor is C)
as i would need to get all three for 2.2, i consequently don't have 2.2.
HLA-DQ7:
rs4639334: not present at all
i think this proves i can't get celiac, except in some obscure cases that nobody currently understands, maybe. maybe. probably not.
but, i cannot source this.
all i can find is abstract language.
but, i think i'm missing a point about the rsid polymorphisms and how they map to gene expressions (alleles). that is, i think that what i'm looking for might be staring me in the face, but that i don't have the mapping to pull it out. but, i can't prove that, either.
1:36
the mapping has to do with these genes: DQA1 and DQB1.
i think that what the results i have are suggesting is that i don't produce specific variants of those genes, and therefore can't produce the immune response.
but, i want an explicit mapping.
2:01
i'm looking for something exceedingly technical and specific, and falling into a black hole where the popular sites are writing it off as too complicated, and the technical sites are writing it off as trivial.
but, surely, somebody wrote the paper.
what was the original celiac paper? let me try that - let me find a history of celiac disease.
2:02
yeah. it's the mapping.
if i had the TT (AA), or the TC/CT (AG) i'd have a dq2.5 cis, heterozygous or homozygous, like in the previously posted study (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4405087/). that would mean i could get celiac. but, i don't - i have the CC (GG), which codes for dq2.5 trans. that means that dqa1*0505 and dqb1*0202 are encoded on different chromosomes and consequently can't be...
so, this paper has the information i need in it:
and i can implicitly deduce the following:
HLA-DQ2.5:
rs2187668: C ----> DQA1*05 [alpha chain] <----i have two of these
rs2187668: T------>DQB1*02 [beta chain]
that would mean TT is the highest risk, TC/CT is moderate risk and CC is zero risk, because you need at least one of DQB1*02. DQA1*05 can come back later, but only in the presence of one of the 2.2 polymorphisms.
so, can i confirm that?
2:44
so, this site seems to present a slightly different mapping, but it doesn't give me the information i want.
it's explicitly suggesting that the A (that is, T - this depends on how you catalog it, as As come with Ts (and Cs come with Gs)) polymorphism is the cause of the "drb" chain, which is more associated with lupus. but, it's enough for me to realize that my deductions appear to be accurate.
i can then check the snpedia page and it reads a little more clearly with that point in mind:
rs2187668(A) also tags the tightly linked DQB1*0201 allele,
so, that tells me that the t polymorphism explicitly codes for the problem allele.
then i can pull this up:
i'm going to post a screenshot of the table, as they're forcing you to download it:
that's it.
it's not that T codes for the dqb and C codes for the dqa, it's that the T codes for them on the same chromosome and the C codes for them on different chromosomes.
so, that's what i've learned here, so far - that rs2187668 CC means that i have dqb1*0202 (not dqb1*0201) on one chromosome and dqa1*0505 on the other. so, i can still get the 2.2 expression, possibly - but i can't get the 2.5 expression.
so, yes - we've ruled out the major form of celiac, which is the combination of dqa1*0505 and dqb1*0201 because:
1) i don't have dqb1*0201, i have dqb1*0202 and
2) dqa1*0505 and dqb1*0202 appear on different chromosomes
it is still possible that the dqa1*0505 could bind to a bad partner, or the dqb1*0202 could bind to a bad partner, but they're not able to bind together.
so, let's move on to the next rsid.
4:42
so, now i'm reading the wiki page on this and it's making sense:
i will still need to worry about the 2.2 expression, because i have the dqb1*0202 allele and will need to see if i have the corresponding dqa1*201 allele, on the same chromosome. if the site i consulted is correct, i should not have them on the same chromosome, and may not have dqa1*201 at all. so, i should probably have the following linkage then: dqa1*303-dqb1*202, and this linkage appears to have no health concerns attached to it, at all - or at least none that i can see at the moment.
can i get as detailed information about rs7454108?
5:03
rs7454108 should map C to the problem allele, DQA1*03/DQB1*0302 and T to some other configuration. let me try to figure out what that is, so i know what configuration i have.
snpedia starts me off:
rs7454108(C) allele is associated with DQB1*0302 (and thus DQ8).
that's pretty clear.
this graphic seems to suggest that rs7454108 does not map to dqb1*0302 unless it is heterozygous, but it doesn't tell me what the T T polymorphism maps to:
but, this seems to be a function of the exact problem. they explain:
The prevalence of the DQB1*0302 allele in Denver is 21.9%. The rs7454108 C allele has a positive predictive value of 97.5% and a negative predictive value of 99.7%. Although we recommend further traditional typing to confirm the presence of high-risk genotypes, the positive predictive value of this two-SNP test is quite high in a relatively low frequency population and demonstrates the utility of this two-SNP test.
The prevalence of the DQB1*0302 allele in Denver is 21.9%. The rs7454108 C allele has a positive predictive value of 97.5% and a negative predictive value of 99.7%. Although we recommend further traditional typing to confirm the presence of high-risk genotypes, the positive predictive value of this two-SNP test is quite high in a relatively low frequency population and demonstrates the utility of this two-SNP test.
so, that chart should really say C/T or C/C - and maybe it's even a typo. hey, do i get paid for peer review? go buy my album.
but, what does T code for, then?
it's not clear.
but it's not hla-dq8 - i don't have that.
5:41
so, i found this:
this is explicit and what i wanted:
1) rs2187668 C C is not dq2.5
2) rs7454108 T T is not dq8
3) rs2395182 T G + rs7775228 T T is not dq2.2
ok.
i'm going to try to do it the long way, anyways.
6:32
so, this is where we're at right now:
1) rs2187668 CC means that i have dqb1*0202 (not dqb1*0201) on one chromosome and dqa1*0505 on the other. therefore, i cannot have hla-dq2.5.
2) rs7454108 TT means i don't have DQB1*0302 and therefore don't have hla-dq8. i can't find more specific information, and would like to, but i have to accept it doesn't matter, in context.
3) what's left is to focus on hla-dq2.2, and i'm going to use the shortcut of rs7775228 T T, which is the important point.
7:34
wait.
can i have two dqa*'s or is the finding on the first dqa actually enough?
8:40
and, what does it mean to say that the hla is on two different chromosomes, as the source did?
i guess that, as we have two copies of chromosome 6, what it's saying is that i have dqb1*0202 on one copy and dqa1*0505 on the other. i mean, how else do you process that?
so, what i'm checking to see is if i have dqa1*202 on the same chromosome (the same copy of chromosome 6) that dqb1*0202 is on, then. well, what i'm trying to figure out is what the dqa1 and dqb1 genes are on the opposite copies of chromosome 6. and, i know i don't have dqb*0302, at all.
but is the homozygosity of the CC actually enough to know what i have for both?
what i know is this:
that says that you get dq2.2 [DQA1*0201-DQB1*0202] 98% of the time when you have the C allele, which i don't have. this table doubles down on the identification, but it's also at wiki.
it doesn't say what likely allele or alleles result from a T allele, or even if the T allele rules out the dqa1*0201.
this table seems to suggest that you don't get dqa1*0201 when at least one of the snps is false, pretty much ever:
but, i'd kind of like something more specific.
hrmmmn.
9:25
so, what do we have then?
where, x≠0201 (rs7775228=TT) and y≠0302 (rs7454108=TT).
we have two copies of chromosome 6, both with hla-dqa1-hla-dqb1 linkages, and it looks like:
| c1 | c2 |
|---|---|
| dqa1*0505 | dqa1*x |
| dqb1*y | dqb1*0202 |
where, x≠0201 (rs7775228=TT) and y≠0302 (rs7454108=TT).
that means, x = 0302 (it's the only remaining option) and y = 0301, according to the following linkage options: https://en.wikipedia.org/wiki/HLA-DQ
so, i have the following:
hla-dq7.5: dqa1*0505-dqb1*0301 [11% frequency]
hla-dq2.3: dqa1*0302-dqb1*0202 [0.08% frequency]
the first haplotype might increase the risk of celiac if hla-dq2.5 is present, but we're into the obscure cases that are probably not celiac, now.
so, i don't have celiac.
like i said.
10:20
hla-dq2.3 seems to be pretty obscure.
but there's a write-up on hla-dq7.5 here:
it seems to be in high proportion amongst middle eastern and italian populations, so i probably got my celiac immunity from my dad. thanks, dad.
also,
DQB1*0301 may be under current positive selection in the human population, at least in areas where DQ2.5 and DQ8 are high, as it confers resistance to type 1 diabetes.
score.
10:59
that's just the cis-pairings, though.
according to the wiki site (https://en.wikipedia.org/wiki/HLA-DQ#Effects_of_heterogeneity_of_isoform_pairing), and basic genetics, i should also have potential trans-pairings, as:
1) dqa1*0505-dqb1*0202 <---that's actually hla-dq2.5 (almost).
2) dqa1*0302-dqb1*0301 <-----that's hla-dq7.3, which is not a celiac risk.
so, what to make of that?
there are apparently obscure cases that might be explained by trans-pairings, but acceptance of this hypothesis is currently a minority position. celiac is a favourite disease of hypochondriacs, and is often misdiagnosed. so, maybe there are rare trans cases, and maybe those cases are actually crohn's or something similar.
i don't have symptoms of celiac, and i'm going to gloss over this obscure point.
you're looking at the obscure of the obscure.
11:36
i think that if i had dq7.5/dq2.2, i'd be at risk for the trans isomer - i think that's why i tested for dq2.2 in the first place.
but, i don't have dq2.2, i have dq2.3. and, that's supposed to neutralize the risk from dq7.5.
i'm sure you'll find somebody, but i do believe that the conventional scientific view is that these genes rule out celiac entirely.
12:01
oct 8, 2021
ok, so that lingering trans isomer wasn't sitting well with me and i couldn't let it go.
i'm now going to go back through the initial post and correct some parts in red.
i think i misread the celiac chart a little - and that the outcome is in my favour.
let's go back to this picture:
i decided that because i had CC in the test, i must have 2.5 trans - because the CC must have a clear outcome. i saw the "or dq8" part but decided it was information related to celiac status, as the snp test was determining one clear outcome or another. and, i appear to have missed the point of what the snp tests are doing, in the process. see, i told you i was missing something obvious.
what i assumed was that the snp tests were a mapping and that each outcome maps to a specific outcome. so, TT goes to one clear outcome, TC goes to another and CC goes to a third. rather, it seems that the snp tests are merely binary chemical tests that could only state more than that by coincidence. so, they're simply testing to see if the T exists or not - and the test only tells you if you have 2.5cis or if you don't.
it follows that if you don't have 2.5cis and you're celiac then you must have 2.5trans or 2.8 in some way. but, it doesn't follow that if you have CC then you have 2.5 trans, and therefore must have 2.8 to explain the celiac status (if you have it).
then, because i was looking for explicit snp mappings rather than realizing that snps are binary tests, i repeatedly went looking for what a negative test would map to and repeatedly got disappointed when i couldn't find it. but, there was nothing to find - negative just means negative. i settled for that with the second two, and constructed a typing via trial and error, but it was based on an unjustified assumption, which left me with an undefined risk status.
so, all i learn from rs2187688 CC is that neither of my 6th chromosomes has dq2.5 on it, in cis. i don't have any further information about what they actually are.
then, going back and looking at the other two, i learn that neither of the beta-chains are dqb1*0302 from rs7454108 TT and that i do not have dq2.2 in cis on either of my 6th chromosomes from rs7775228 T T.
that leaves me with a large number of potential outcomes, as the following charts, placed side by side, indicate:
so, i had previously decided that i had the trans-isomer and it was of minimal concern. in fact, i don't know what allele i have at all, i just know i don't have the cis isomers for the above three gene expressions on either chromosome, because the three snps are all homozygous. so, i'm deciding that the widespread typing as seen on the internet is incomplete and i want to find an snp that tests for the trans isomer before i decide that i'm completely risk free.
listen - that was a first run, and the fact that it didn't sit right with me and i realized there was something wrong is a good thing.
i'll get this corrected soon.
5:25
this graphic demonstrates the point at hand.
(graphic from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4333989/)
you can see in the graphic that a couple of cases were found in the α5 and β2 chains in ways that were disconnected from the the three risk factors of hla-dq2.5, hla-dq2.2 and hla-dq8:
but, you'll also note that the p-values suggest the association isn't statistically valuable.
the risk factor, however dubious, would be associated with the hla-dq7.5/hla-dq2.3 that i erroneously deduced that i had. rather, that would be the worst case scenario - where i would still have a dubious risk from the cross factor.
hey, it's been a long time since first year bio, but it's clear now. it would take most people more than a week to figure this out in the detail i'm demanding understanding it in.
the extra snp - which is missing in my data - would seek to rule out the dq7, which is the α5 cross-factor. but, it seems like it's more valuable to me to try to rule out the β2 cross-factor.
regardless, let's see what i can do about this.
6:08
i am at least confident that i understand how this works, now.
but you'd think it wouldn't be so annoying to simply find a list of snps to search for, to understand if you have the remnant types. a complete typology shouldn't be that hard.
i also would like to know what it means, exactly, when an snp is not present, and am not sure why that's so hard, either.
8:05
ok, so i think this is what i wanted to find:
but, the source is not good.
so, now i'm trying to prove or disprove this.
8:39
here's some results:
hla-dqa*0101:
rs8365: C G <---risk variant is C
rs482044: G C <------risk variant C
rs532098 AC <-----risk variant C
===============================
i have hla-dqa*0101 ======> i'm hla-dq5.1 in cis on one chromosome
hla-dqa*0102:
rs5875382: not present
rs1063355: not present
hla-dqa*0103:
rs7744001: not present
rs1063355: not present
rs2187686: not present
hla-dqa*0201:
rs7763805: not present
rs384431:not present
hla-dqa*0301:
rs2395533: not present
rs6903433: C C <----risk allele is C
hla-dqa*0401:
rs1694112: not present
rs4711312: not present
hla-dqa*0501:
rs4639334:not present
rs6908943: not present
hla-dqa*0601:
rs1061172: not present
rs312971: not present
well, that's one potential answer.
let's try the beta - and remember that these are coming from an unreliable source, so i need to verify them.
hla-dqb*0201:
rs4988889: not present
hla-dqb*0301:
rs6928482: not present
rs1056315: not present
hla-dqb*0302
rs6906021: T T <----risk factor is T.
rs9275184: not present
hla-dqb*0303
rs2395533: not present
rs2856683: not present
hla-dqb*0401
rs2077580: A C <----risk factor is C
rs2395533: not present
rs2300825: not present
hla-dqb*0402:
rs2076528: not present
rs1694112: not present
rs6937034:not present
hla-dqb*0501:
rs4947332: C C <----risk factor is C
====================================
this is the second part of hla-dq5.1. is it homozygous, though?
hla-dqb*0502:
rs7744001: not present
rs6906021: T T <----risk factor is T.
rs2300825: not present
hla-dqb*0503:
rs1794265: not present
rs241446: C C <---risk factor is C
hla-dqb*0601:
rs7744001: not present
rs6928482: not present
hla-dqb*0602:
rs3134975: not present
rs2621426: not present
hla-dqb*0603:
rs3806155: not present
hla-dqb*0604:
rs11758998: not present
that's potentially one clear hit.
let's see if i can make sense of that.
9:25
rs4947332 seems to be a tag for an hla-drb gene that is only correlated with the hla-dqb1*0501 allele. it's in the right neighbourhood of the 6th chromosome, but i'm only getting weak results. the hits want to present a different snp i can't find.
let me try these ones instead:
hla-dqa*0101:
rs8365: C G <---risk variant is C
rs482044: G C <------risk variant C
rs532098 AC <-----risk variant C
9:50
ok, i see.
these tags are population specific - i got a chinese population. it was indeed from a chinese blog.
but, they don't carry on outside of chinese populations. it's strictly correlational.
that said, i found a list with european populations and it might give me more of a hint:
10:27
i'm getting some leads in studying narcolepsy, and it would seem to suggest that i'm hla-drb1*15:01-dqb1*0602.
but i need to stop to eat.
10:45
ok, i'm willing to bite on this. these are very different articles, but the idea is that, if i show i have alleles that aren't celiac, i've shown i don't have alleles that are.
so, let's look at a few entirely unrelated studies.
first, i suppose we should just put aside the bit about being at higher risk for ms (a point i've long suspected, regardless):
i'm going to need to deal with the ms thing on it's own.
for now, note that rs3135388-T is a tagging snp for dqb1*0602 - and i have that. it's T/C, to be more precise.
but, that's just one source and that's not good enough.
that study is about a drug that produces liver damage, but it pulls out the same tagging snp.
the line of thought that brought me down this path had to do with studying the effects if dqb1*0602 on narcolepsy, so it's perhaps fitting that i'm going to pass out.
but, we have this at least - one of my chromosomes has dqb1*0602 on it, which allows for one of the following heterodimers, in cis: dq6.1 or dq6.2.
12:57
so everybody cites "de bakker et al", but de bakker et al don't show their work.
ugh.
the master document would appear to be table 3 from this article:
but, that's not good enough for me.
14:29
yeah, bakker et al (2005) seem to have used an algorithm based on least-squares. they just dump it out in bakker et al (2006), and they even point out "but, this might not be right, guys".
i want to back each point up with an explicit empirical study, not a dart-throwing least squares maybe-guess.
so, bakker et al (2005/2006) is a starting point, not an ending point.
ok - now i grasp that.
14:44
you upset?
yeah?
fuck bakker et al.
14:47
oct 9, 2021
ok.
so, the least squares guesses keep throwing hla-dr* snps at me, and i keep throwing them away.
but, i'm realizing that some of the hla-dr* typing links uniquely to specific hla-dq serotypes.
so, there may have been better answers there than i thought.
11:47
so, rs3135388-T is really a tag for the hla-drb1*1501 and the claim in those articles was that hla-drb1*1501 links uniquely to hla-dqb1*0602.
the wiki site (https://en.wikipedia.org/wiki/HLA-DR15) confirms the association to hla-dq6 and is specific about the dqa1 allele, as well.
so, i think i can be more explicit - rs3135388-T codes for dqa1*0102-dqb1*0602, which is hla-dq6.2, in cis.
so, that's the result on one chromosome, and it is clear - which gives me no risk for celiac on that chromosome, in cis - but a higher risk for ms. i can't get cervical cancer. it's a substring of this bigger gene:
one will note that none of the cis celiac risk alleles (2.2, 2.5, 8.2 or the questionable 9) and none of the questionable trans risk alleles (2.3, 4.3, 7*, 9*) have dqa1*01 or dqb1*06, so none of them could be formed from any cross pairing.
so, is that enough?
the following alleles, which could still exist on the other chromosome, remain obscure risks:
- 2.3 [due to the beta chain]
- 7.5 [due to the alpha chain]
if we're going to do this, let's do this until it's done.
12:25
to summarize, so far:
rs2187668 = C C ----> not(hla-dq2.2, which is dqa1*0501-dqb1*0201) (in cis)
rs7454108 = T T ----> not(hla-dqb1*0302) on either chromosome
rs7775228 = T T -----> not (hla-dq2.2, which is dqa1*0201-dqb1*0202) (in cis)
and
rs3135388-T (from T C) -----> hla-dq6.2, which is dqa1*0102-dqb1*0602,
12:35
so, the first thing that's coming up is that tentative least-squares association of rs14004 (C/G) with dqa1*0102 - which is the first part of hla-dq6.2.
i'm wondering if the tag, being homozygous, is enough to deduce i also have dqa1*0102 on the other chromosome.
but, i can't get there.
at the least, it's not surprising that it came up. let's put that aside.
12:57
rs660895 comes up as T T, which means i don't have drb1*0401 (https://www.snpedia.com/index.php/Rs660895), which links to
- dqa1*0301 and dqa1*0303
- dqb1*0301 and dqb1*0302 (rs7454108)
if i could show that not having drb1*0401 means not having dqa1*0301 or dqa1*0303, i would disprove some variants.
however, the list at wiki (https://en.wikipedia.org/wiki/HLA-DQ4#DQ4_distribution) shows that:
1) dqa1*0303 is frequently linked to drb1*0405 through the plausible dqb1*0401
2) dqb1*0301 links to multiple drb1* alleles through hla-dq7*, but none of them to dqa1*0301.
further, dqa1*0301 links readily to drb1*0402 and drb1*0404, but only through the discarded dqb1*0302 (and hla-dq8). so, can i disprove dqa1*0301 by disproving drb1*0401, given that i have already disproven hla-dq8?
actually, let's look at that chart a little more closely:
- i don't have drb1*0401, by the above snp
- i don't have dqb1*0302, so i don't have drb1*0402 or drb1*0404
if i have hla-dq4 at all, it would have to be dqa1*0303-dqb1*0401-drb1*0405.
and, this seems to be a dead-end.
13:45
ok, so the other tag i have is rs2858880-T. in fact, i have T T.
this study clarifies:
that's a very minor allele with a very strong correlation.
this study had the same findings:
going back to the previously posted jpg at wiki:
drb*1502 has only one linkage - hla-dq6.1.
so, am i hla-dq6.1/hla-dq6.2? or am i misinterpreting something?
well, i've tred to search for rs285880 as a tag for drb*1501 and nothing has come up. and, i've likewise searched the previous snp (rs3135388) for drb*1502 and nothing came up. on the other hand, bakker et al separates them, and the testing is very specific.
that doesn't mean it isn't a false positive, but i have no evidence that it is.
but, there's more.
17:16
if i go through the list of suggestions in bakker et al, these are the least squares guesses, how many of the options (for the ceu) actually produce correct matches?
rs6457594 does not exist <-----tag is T
rs2157051 does not exist <-----tag is C
there's three - and no more:
hla-dqb1*0601: rs2858880 TT <----tag is T
hla-dqb1*0602: rs3135388 CT <-----tag is C
hla-dqb1*0603: rs2395150 TC <------tag is T
rs3135461 TC <------tag is C
yes - all in the *06 space, which is strongly telling. but, i only have two, so, what are the right answers?
let's look at drb.
hla-drb1*1501: rs3135388 CT <------tag is C
hla-drb1*1502: rs2858880 TT <----tag is T
hla-drb1*1301: rs2395173 TT <----tag is C
rs2157051 does not exist <------tag is C
lastly, let's look at the corresponding dqa1* linkages:
hla-dqa1*0102: rs14004 CC <----tag is C
hla-dqa1*0103: rs2273017 T C <----tag is T
now, let's look at the three potential haplotypes:
1) drb1*1501-dqa1*0102-dqb1*0602
2) drb1*1502-dqa1*0103-dqb1*0601
3) drb1*1301-dqa1*0103-dqb1*0603
so, tagging these independently isn't specific enough. while it may turn out to be an error, i'm going to present the following typing, for the exceedingly rare case that somebody may be homozygous in dq6, and move on:
1) drb1*1501-dqa1*0102-dqb1*0602:
- rs3135388 c & rs14004 C
2) drb1*1502-dqa1*0103-dqb1*0601
- rs2858880 T & rs2273017 T
3) drb1*1301-dqa1*0103-dqb1*0603
- rs2395173 T & rs2395150 T & rs3135461 C & rs2273017 T
that said, i should point out that i also tagged positively for dqa*0101 in the chinese population, but couldn't back up any of the correlations.
worse, hla-dq6.1 is exceedingly rare in western populations and hla-dq6.3 is only sort of rare.
so, the total answers here may be somewhat blurry. i may be missing tagging snps, and i may even have a weird combination. like, maybe i'm dqa1*0101-dqb*0603 on one chromosome, or something.
but, the basic point is clear enough, i think - i'm homozygous in dq6.
18:05
this result is unusual, but it is what it is.
it's not impossible that something went wrong during meiosis:
18:23
i'll run through that again in a bit.
18:23
so, there's no chance of celiac and no chance of diabetes, but a very high risk factor for ms.
i need to make sure my d's high and stay away from the smoke, then.
but, i think the better way to look at it is that i'm homozygous in hla-drb1*15 - that's what the snps really say.
18:31
am i immune to aids?
i'm not doing that experiment.
sorry.
18:33
oddly, dq6.1 seem to protect against ms - as dq6.2 is the strongest risk factor for it.
hrmmn.
18:41
and, then there's this: